1、使用SimpleITK对齐图像
在看voxelmorph的代码,看到图像对齐部分,记录一下。
下面是从voxelmorph项目中截取的一段保存图像的函数。
函数输入分别是:配准后的图像、固定图像、要将配准图像保存的名字。
将图像对齐的操作需要将对齐的图像的原点、方向、间距设置成与 被对齐的图像一致。
1 def save_image(img, ref_img, name):
2 img = sitk.GetImageFromArray(img[0, 0, ...].cpu().detach().numpy())
3 img.SetOrigin(ref_img.GetOrigin())
4 img.SetDirection(ref_img.GetDirection())
5 img.SetSpacing(ref_img.GetSpacing())
6 sitk.WriteImage(img, os.path.join(args.result_dir, name))
这里只是做了属性的修改,并没有真正的对齐。
2、重采样并对齐图像
1 def align_seg_with_raw_nrrd(dcm, seg):
2 # Just for labelmap .... because of nearestNeighour interpolator
3 resampler = sitk.ResampleImageFilter()
4 resampler.SetReferenceImage(dcm)
5 resampler.SetTransform(sitk.Transform(3, sitk.sitkIdentity))
6 resampler.SetInterpolator(sitk.sitkNearestNeighbor)
7 seg_new = resampler.Execute(seg)
8 return seg_new
9
10 if __name__ == '__main__':
11 raw_dcm_file = r"path to your nrrd raw data"
12 seg_dcm_file= r"path to your seg file"
13 dcm = sitk.ReadImage(raw_dcm_file)
14 seg = sitk.ReadImage(seg_dcm_file)
15 print(raw_dcm_file, seg_new_file)
16 seg_new = align_seg_with_raw_nrrd(dcm, seg)
我要实现的目的是:将原始stl切下与原始数据重合的部分,并且用一个与原始数据一样的尺寸存放,并且对应的空间坐标一致(原点、spacing、direction)。
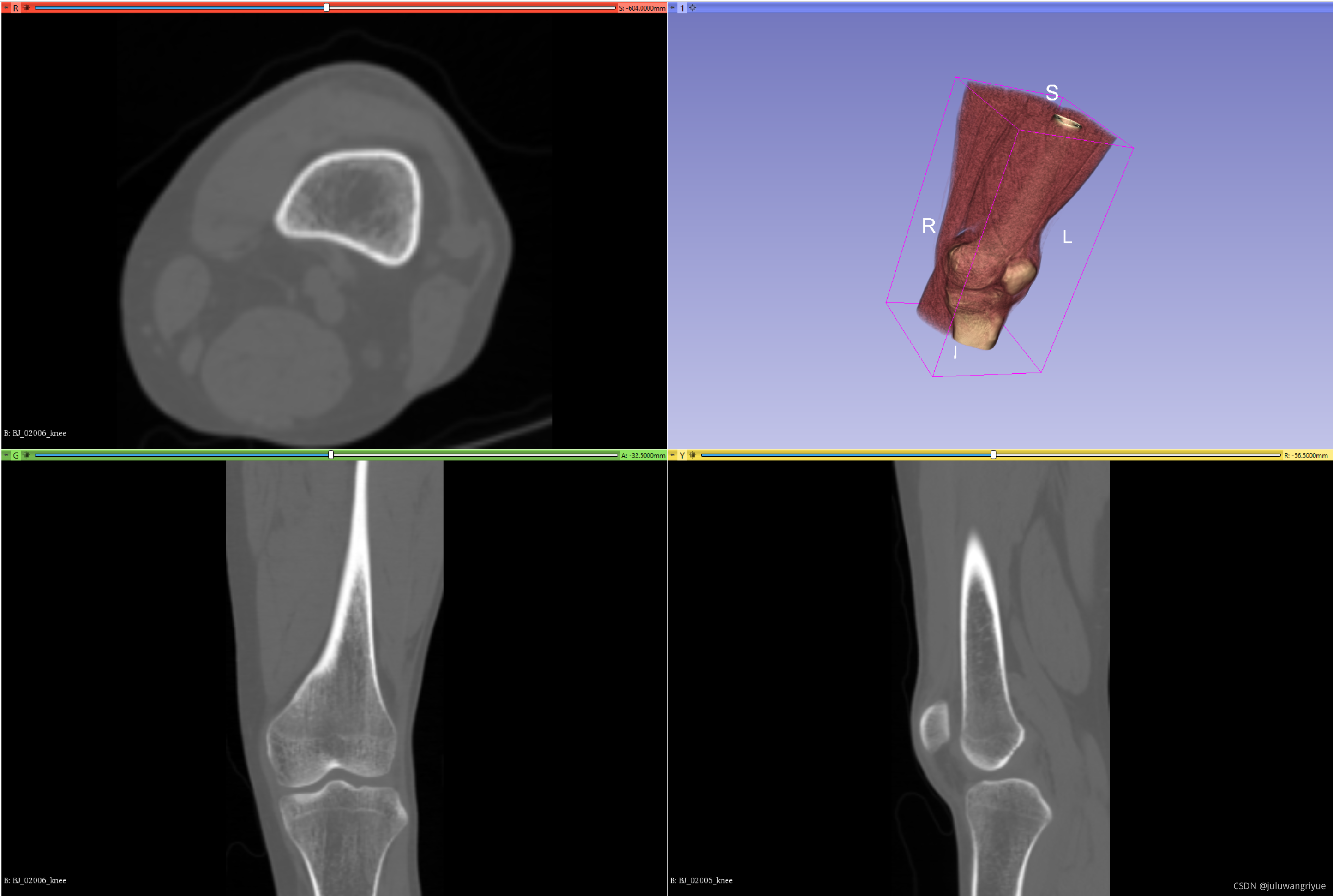
如下输入原始数据:

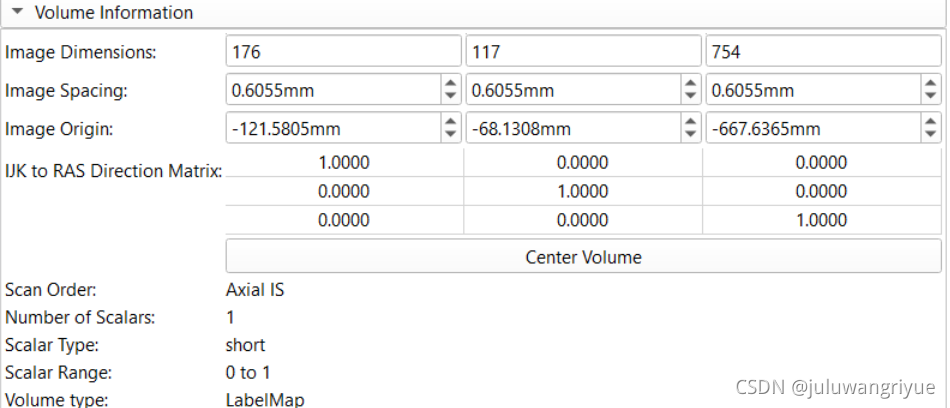
原始的stl数据,绿色部分:
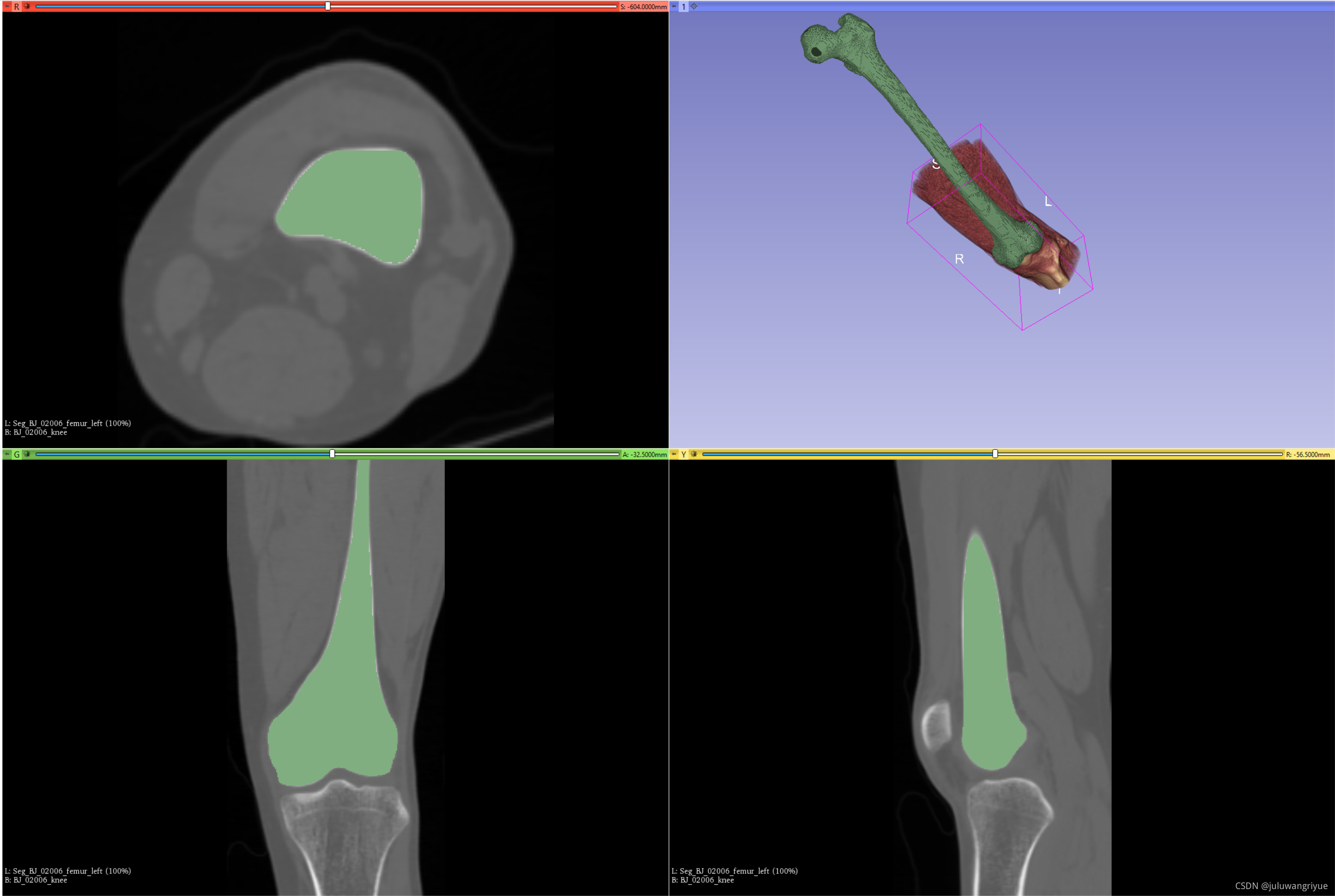
对齐后的效果如下:
stl对齐后的volume信息如下
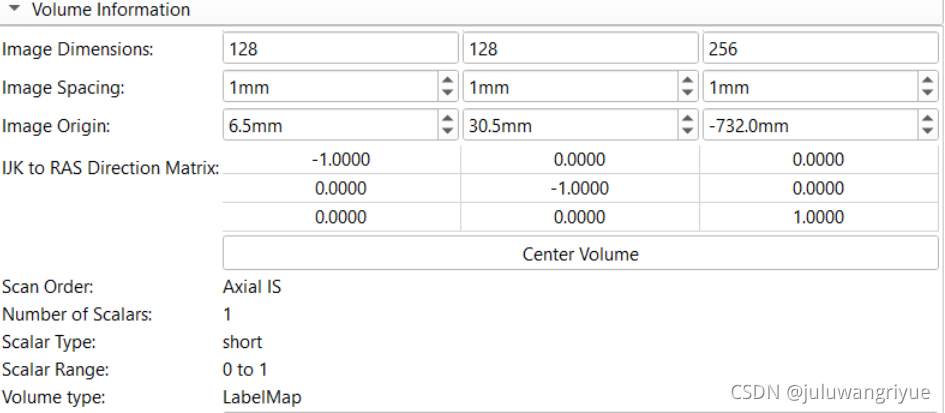
对齐后的渲染效果如下

3、中间走过的弯路
多设置参数,得到错误的结果。
4、尺寸不一致时改变方向并对齐
1 # -*- coding : UTF-8 -*-
2 # @file : resample_seg.py
3 # @Time : 2021/10/14 0014 19:58
4 # @Author : wmz
5
6 import os
7 import SimpleITK as sitk
8 from glob import glob
9 import numpy as np
10
11
12 def chnm_stdraw2seglabel(stdrawfile):
13 seglabelfile = stdrawfile
14 seglabelfile = seglabelfile.replace("_knee", "")
15 if right_flag:
16 if femur_flag:
17 seglabelfile = 'Seg_' + seglabelfile[:-5] + '_femur_right.nrrd'
18 else:
19 seglabelfile = 'Seg_' + seglabelfile[:-5] + '_hip_right.nrrd'
20 else:
21 if femur_flag:
22 seglabelfile = 'Seg_' + seglabelfile[:-5] + '_femur_left.nrrd'
23 else:
24 seglabelfile = 'Seg_' + seglabelfile[:-5] + '_hip_left.nrrd'
25 return seglabelfile
26
27
28 def align_seg_with_raw_dcm(dcm, seg):
29 # Just for labelmap .... because of nearestNeighour interpolator
30 # 读取文件的size和spacing信息
31 outsize = [0, 0, 0]
32 inputsize = seg.GetSize()
33 inputspacing = seg.GetSpacing()
34 input_origin = seg.GetOrigin()
35 dcm_size = dcm.GetSize()
36 dcm_spacing = dcm.GetSpacing()
37 dcm_origin = dcm.GetOrigin()
38 direction = dcm.GetDirection()
39 seg_direction = seg.GetDirection()
40
41 transform = sitk.Transform()
42 transform.SetIdentity()
43 # 计算改变spacing后的size,用物理尺寸/体素的大小
44 outsize[0] = round(inputsize[0] * inputspacing[0] / dcm_spacing[0])
45 outsize[1] = round(inputsize[1] * inputspacing[1] / dcm_spacing[1])
46 # outsize[2] = round(inputsize[2] * inputspacing[2] / dcm_spacing[2]) # 原大尺寸,弃用
47 outsize[2] = round((dcm_size[2] + dcm_origin[2] - input_origin[2])/dcm_spacing[2])
48
49 resampler = sitk.ResampleImageFilter()
50 resampler.SetReferenceImage(dcm)
51 resampler.SetTransform(sitk.Transform(3, sitk.sitkIdentity))
52 resampler.SetInterpolator(sitk.sitkNearestNeighbor)
53 resampler.SetOutputOrigin(seg.GetOrigin())
54 resampler.SetOutputSpacing(dcm.GetSpacing())
55 resampler.SetOutputDirection(seg.GetDirection())
56 resampler.SetSize(outsize)
57 seg_new = resampler.Execute(seg)
58 return seg_new
59
60
61 def align_label2stdraw(stdrawfile, labelfile):
62 std_img = sitk.ReadImage(stdrawfile)
63 std_img_size = std_img.GetSize()
64 std_img_direction = std_img.GetDirection()
65 label_img = sitk.ReadImage(labelfile)
66 new_seg = align_seg_with_raw_dcm(std_img, label_img)
67 new_seg_origin = new_seg.GetOrigin()
68 new_seg_direct = new_seg.GetDirection()
69 new_seg_spacing = new_seg.GetSpacing()
70 new_seg_size = new_seg.GetSize()
71 new_seg_data = sitk.GetArrayFromImage(new_seg)
72 new_seg_data = np.flip(new_seg_data, 1)
73 new_seg_data = np.flip(new_seg_data, 2)
74 stdlabel_img = sitk.GetImageFromArray(new_seg_data)
75 flip_seg_origin = [new_seg_origin[0] + new_seg_direct[0]*new_seg_spacing[0]*new_seg_size[0],
76 new_seg_origin[1] + new_seg_direct[4]*new_seg_spacing[1]*new_seg_size[1], new_seg_origin[2]]
77 stdlabel_img.SetOrigin(flip_seg_origin)
78 stdlabel_img.SetSpacing(new_seg_spacing)
79 stdlabel_img.SetDirection(std_img_direction)
80
81 name = labelfile.split("\\")[-1]
82 sitk.WriteImage(stdlabel_img, os.path.join(dst_dir, name))
83
84
85 if __name__ == '__main__':
86 std_full_dir = r'F:\dataset\align_data\std_raw'
87 label_dir = r"F:\dataset\align_data\Seg_volume"
88 dst_dir = r"F:\dataset\align_data\output"
89 right_flag = True
90 femur_flag = False
91
92 files = glob(std_full_dir + '/*.nrrd')
93 for indx, file in enumerate(files):
94 print("processing", indx+1, "of", len(files))
95 print("processing file:", file)
96 # file = r"D:\Work\Data\Patient_Data\train_data\knee_femur\left\NanFang_03021_knee.nrrd"
97 std_raw_file = file.split('\\')[-1]
98 seg_label_file = chnm_stdraw2seglabel(std_raw_file)
99 org_label = os.path.join(label_dir, seg_label_file)
100 if not os.path.exists(org_label):
101 print(org_label, "file not exist !")
102 continue
103 align_label2stdraw(file, org_label)
关键部分是下面这几句:
1 new_seg_data = np.flip(new_seg_data, 1)
2 new_seg_data = np.flip(new_seg_data, 2)
3 stdlabel_img = sitk.GetImageFromArray(new_seg_data)
4 flip_seg_origin = [new_seg_origin[0] + new_seg_direct[0]*new_seg_spacing[0]*new_seg_size[0],
5 new_seg_origin[1] + new_seg_direct[4]*new_seg_spacing[1]*new_seg_size[1], new_seg_origin[2]]
6 stdlabel_img.SetOrigin(flip_seg_origin)
7 stdlabel_img.SetSpacing(new_seg_spacing)
8 stdlabel_img.SetDirection(std_img_direction)